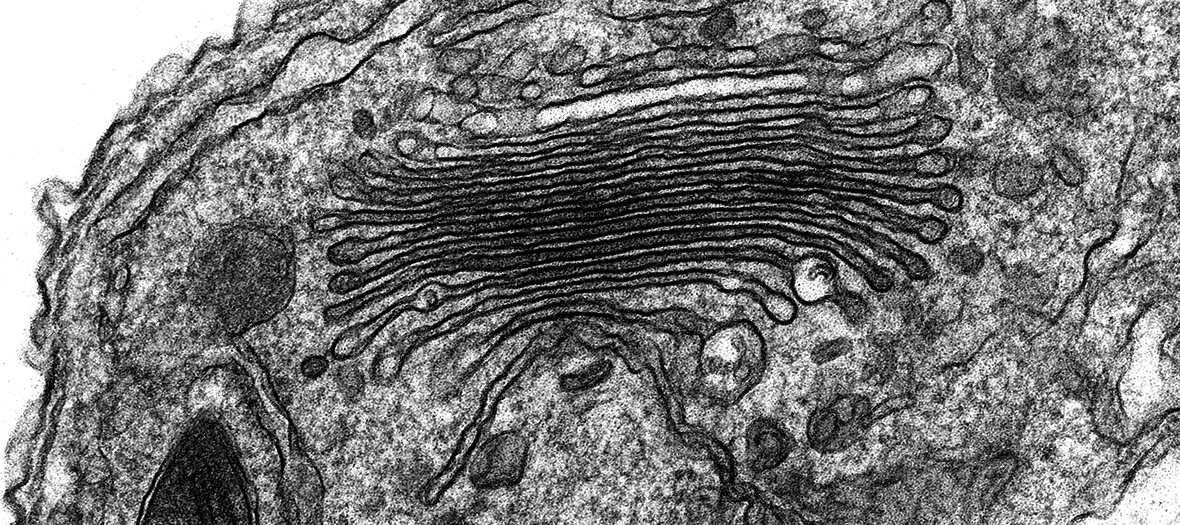
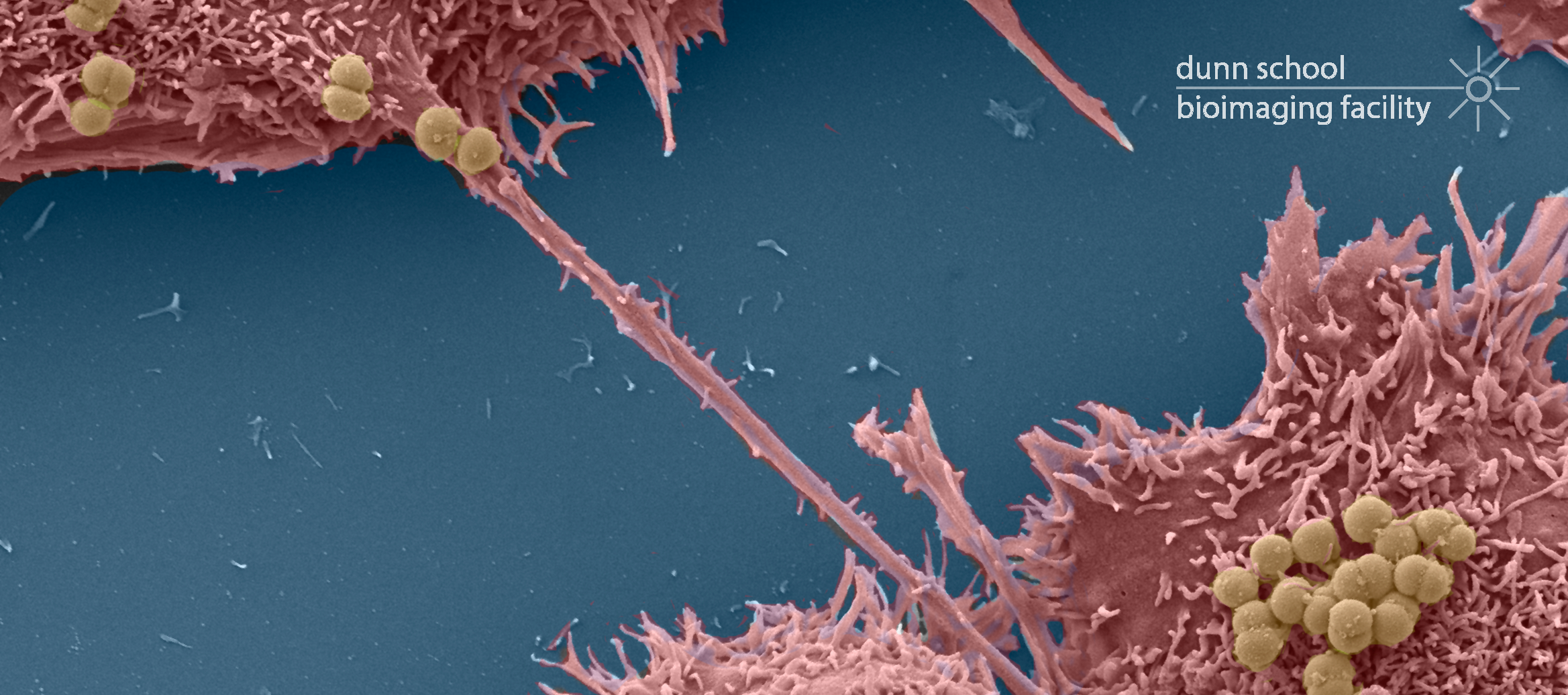
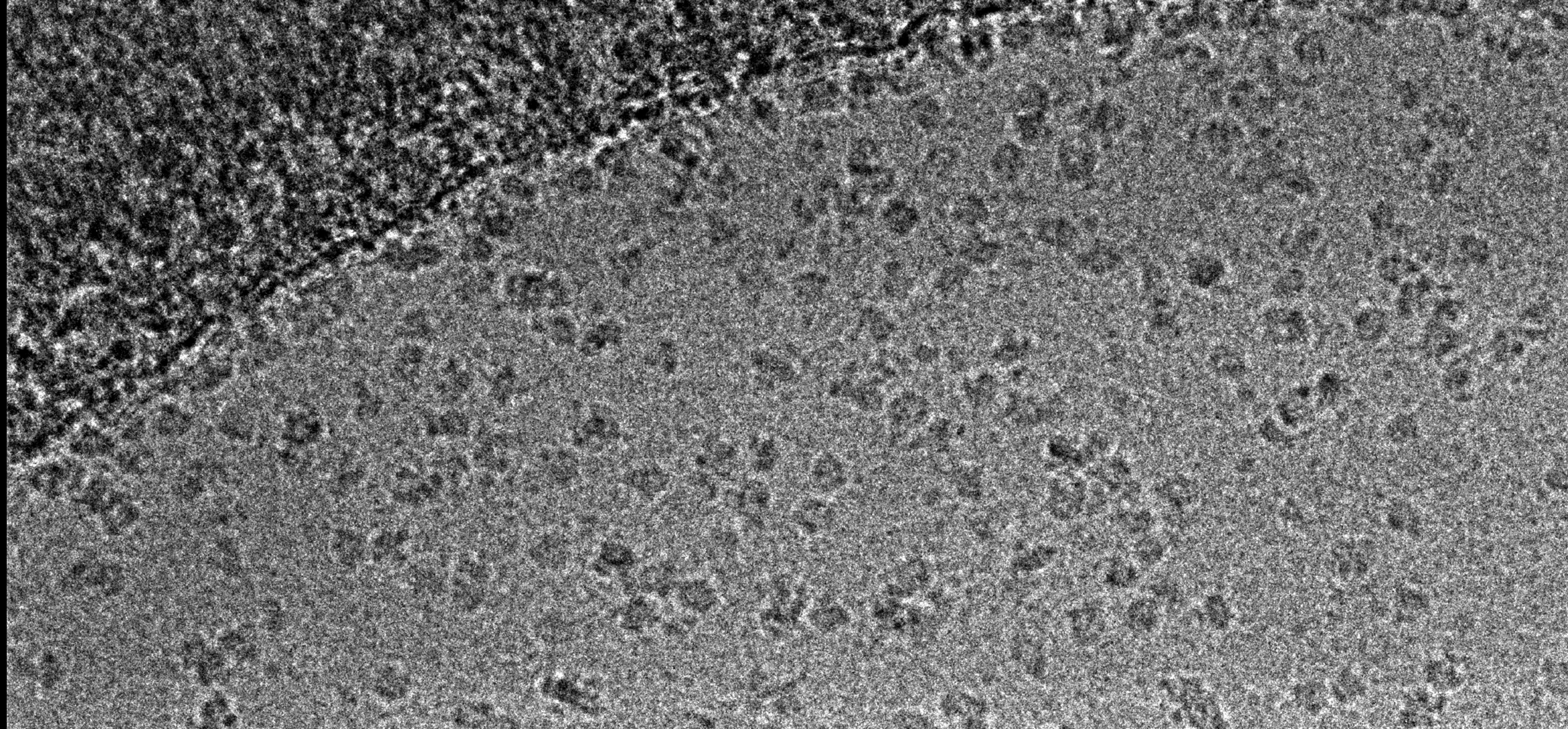
Transmission Electron Microscope (TEM)
Our brand new suite of JEOL TEMs was installed early 2023. These microscopes are more user friendly and higher resolution than our old T12 TEM. They are equipped with high resolution, ultra-fast CMOS cameras to help you get the best ultrastructural images possible. In particular, the JEOL 2100Plus dramatically expands our 3D imaging capabilities at the organelle and cellular levels. This microscope enables higher resolution EM tomography, as well as STEM tomography, to be performed on thicker resin sections and also introduces TEM array tomography capability to our facility, which is very exciting!
Specifications JEOL 1400 TEM:
- Accelerating voltage: 80-120kV
- Filament: Tungsten
- Camera: Gatan Rio CMOS detector
- Resolution: 0.2nm
- Magnification: ×10 to ×1,200,000
- Intended use: Workhorse TEM (similar to our old Tecnai T12), very user friendly
Specifications JEOL 2100Plus TEM:
- Accelerating voltage: 80-200kV
- Filament: LaB6
- Camera: Gatan OneView detector
- Resolution: 0.14 nm
- Magnification: x30 to x8,000,000
- Extra features: Scanning Transmission Electron Microscope (STEM) mode for tomography of thick resin sections
- Intended use: Higher end TEM for more advanced imaging (eg: tomography, negative stain Single Particle Analysis)
Applications:
- Screening negatively stained particulate samples (eg: viruses, bacteria, protein and liposomes)
- Characterisation of cellular ultrastructure (eg: mitochondria, centrosomes etc)
- Subcellular localisation of proteins using immunogold labelling
- Correlative light and electron microscopy (CLEM) to place fluorescent proteins/markers in ultrastructural context
- Electron tomography for high resolution 3D reconstructions of organelles and particulate samples, including using STEM.
Recommended reading:
- Carter, C.B. and Williams, D.B., 2009. Transmission electron microscopy. Springer-Verlag US.
- Hayat, M.A., 1981. Principles and techniques of electron microscopy. Biological applications. Edward Arnold.
- Bozzola, J.J. and Russell, L.D., 1999. Electron microscopy: principles and techniques for biologists. Jones & Bartlett Learning.
- Pavelka, M. and Roth, J., 2015. Functional ultrastructure: atlas of tissue biology and pathology. Springer.
- Harris, J.R., 1991. The negative staining-carbon film procedure: technical considerations and a survey of macromolecular applications. Micron and microscopica acta, 22(4), pp.341-359.
- De Boer, P., Hoogenboom, J.P. and Giepmans, B.N., 2015. Correlated light and electron microscopy: ultrastructure lights up!. Nature methods, 12(6), pp.503-513.
- McIntosh, R., Nicastro, D. and Mastronarde, D., 2005. New views of cells in 3D: an introduction to electron tomography. Trends in cell biology, 15(1), pp.43-51.
Selected recent publications from the Dunn School EM Facility (TEM):
- Goolam, M., Xypolita, M.E., Costello, I., Lydon, J.P., DeMayo, F.J., Bikoff, E.K., Robertson, E.J. and Mould, A.W., 2020. The transcriptional repressor Blimp1/PRDM1 regulates the maternal decidual response in mice. Nature Communications, 11(1), pp.1-12.
- Ruan, J. L., Browning, R. J., Yildiz, Y. O., Bau, L., Kamila, S., Gray, M. D., & Stride, E. (2021). Evaluation of Loading Strategies to Improve Tumor Uptake of Gemcitabine in a Murine Orthotopic Bladder Cancer Model Using Ultrasound and Microbubbles. Ultrasound in Medicine & Biology.
- Scott, C., Downes, D.J., Brown, J.M., Beagrie, R., Olijnik, A.A., Gosden, M., Schwessinger, R., Fisher, C.A., Rose, A., Ferguson, D.J. and Johnson, E., 2020. Recapitulation of erythropoiesis in congenital dyserythropoietic anaemia type I (CDA-I) identifies defects in differentiation and nucleolar abnormalities. Haematologica.
- Hall-Roberts, H., Agarwal, D., Obst, J., Smith, T.B., Monzón-Sandoval, J., Di Daniel, E., Webber, C., James, W.S., Mead, E., Davis, J.B. and Cowley, S.A., 2020. TREM2 Alzheimer’s variant R47H causes similar transcriptional dysregulation to knockout, yet only subtle functional phenotypes in human iPSC-derived macrophages. Alzheimer's research & therapy, 12(1), pp.1-27.
- Davies, A., Kim, H., Gonzalez-Cano, R., Choi, J., Back, S., Roh, S., Johnson, E., Gabriac, M., Kim, M-S., and Lee, J., 2019. Natural killer cells degenerate intact sensory afferents following nerve injury. Cell, in press.
- Riffelmacher, T., Clarke, A., Richter, F.C., Stranks, A., Pandey, S., Danielli, S., Hublitz, P., Yu, Z., Johnson, E., Schwerd, T. and McCullagh, J., 2017. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity, 47(3), pp.466-480.
- Cervellini, I., Galino, J., Zhu, N., Allen, S., Birchmeier, C. and Bennett, D.L., 2018. Sustained MAPK/ERK activation in adult Schwann cells impairs nerve repair. Journal of Neuroscience, 38(3), pp.679-690.
- Little, C.A., Batchelor-McAuley, C., Young, N.P. and Compton, R.G., 2018. Shape and size of non-spherical silver nanoparticles: implications for calculating nanoparticle number concentrations. Nanoscale, 10(34), pp.15943-15947.
- Guidi, L.G., Holloway, Z.G., Arnoult, C., Ray, P.F., Monaco, A.P., Molnár, Z. and Velayos-Baeza, A., 2018. AU040320 deficiency leads to disruption of acrosome biogenesis and infertility in homozygous mutant mice. Scientific Reports, 8.
- Wang, S., Idrissi, F.Z., Hermansson, M., Grippa, A., Ejsing, C.S. and Carvalho, P., 2018. Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nature Communications, 9(1), p.2939.
- Russell, R.A., Chojnacki, J., Jones, D.M., Johnson, E., Do, T., Eggeling, C., Padilla-Parra, S. and Sattentau, Q.J., 2017. Astrocytes Resist HIV-1 Fusion but Engulf Infected Macrophage Material. Cell Reports, 18(6), pp.1473-1483.
- Song, L., Able, S., Johnson, E. and Vallis, K.A., 2017. Accumulation of 111In-Labelled EGF-Au-PEG Nanoparticles in EGFR-Positive Tumours is Enhanced by Coadministration of Targeting Ligand. Nanotheranostics, 1 (3), 232-243.
- Cottee, M.A., Muschalik, N., Johnson, S., Leveson, J., Raff, J.W. and Lea, S.M., 2015. The homo-oligomerisation of both Sas-6 and Ana2 is required for efficient centriole assembly in flies. Elife, 4, p.e07236.
- Johnson, E., Seiradake, E., Jones, E.Y., Davis, I., Grünewald, K. and Kaufmann, R., 2015. Correlative in-resin super-resolution and electron microscopy using standard fluorescent proteins. Scientific reports, 5.
- Brady, J.P., Claridge, J.K., Smith, P.G. and Schnell, J.R., 2015. A conserved amphipathic helix is required for membrane tubule formation by Yop1p. Proceedings of the National Academy of Sciences, 112(7), pp.E639-E648.
- Brune, K.D., Leneghan, D.B., Brian, I.J., Ishizuka, A.S., Bachmann, M.F., Draper, S.J., Biswas, S. and Howarth, M., 2016. Plug-and-Display: decoration of Virus-Like Particles via isopeptide bonds for modular immunization. Scientific Reports, 6, p.19234.
- Pratt, M.B., Titlow, J.S., Davis, I., Barker, A.R., Dawe, H.R., Raff, J.W. and Roque, H., 2016. Drosophila sensory cilia lacking MKS proteins exhibit striking defects in development but only subtle defects in adults. J Cell Sci, 129(20), pp.3732-3743.
- Serpell, C.J., Rutte, R.N., Geraki, K., Pach, E., Martincic, M., Kierkowicz, M., De Munari, S., Wals, K., Raj, R., Ballesteros, B. and Tobias, G., 2016. Carbon nanotubes allow capture of krypton, barium and lead for multichannel biological X-ray fluorescence imaging. Nature Communications, 7.
1400 TEM
2100 TEM