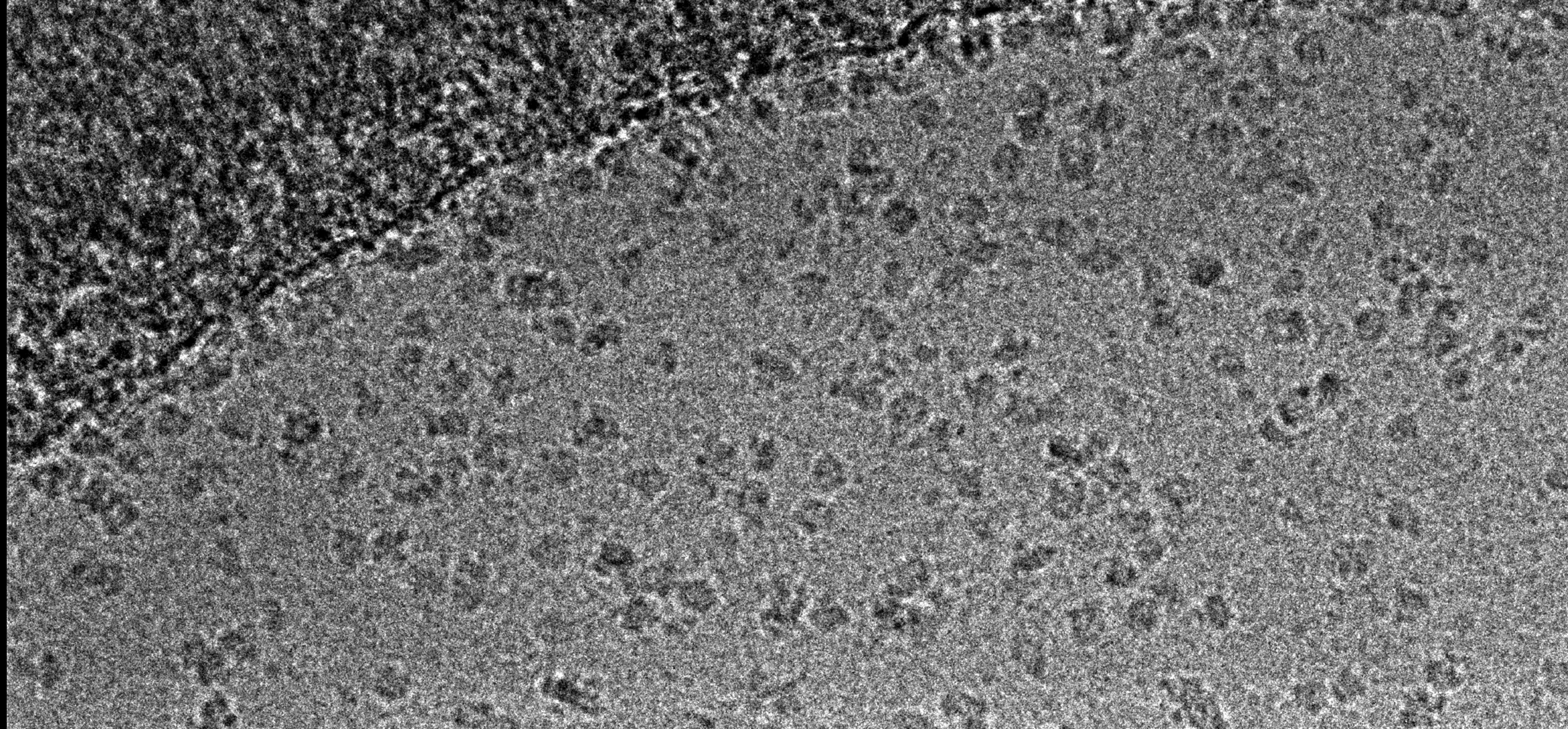
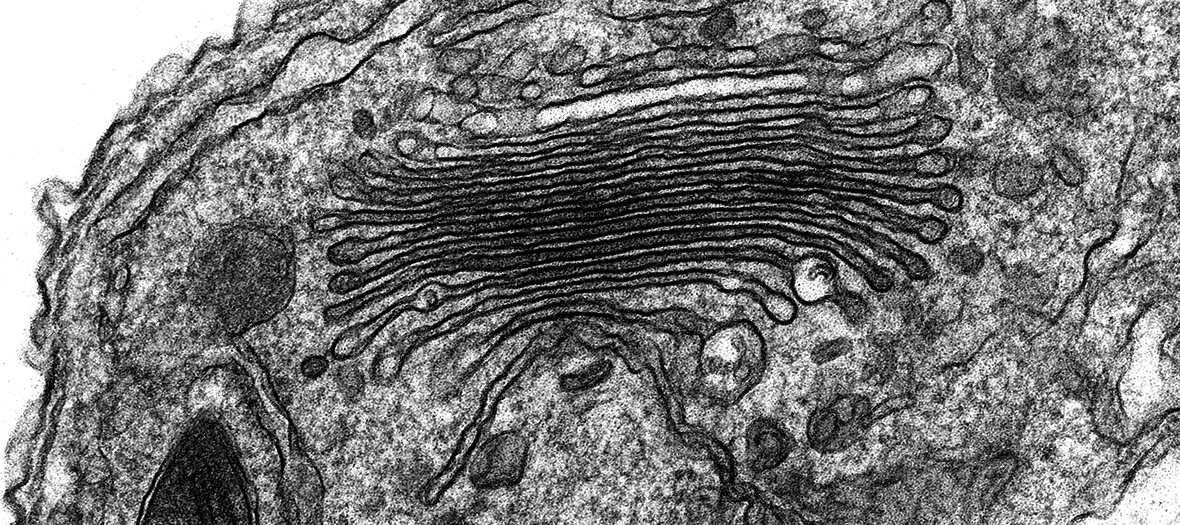
Cryo-electron microscopy
The Central Oxford Structural Molecular Imaging Centre (COSMIC) is a state-of-the-art cryo-electron microscopy facility in the South Parks Rd Science Area at Oxford University. Launched in 2016 in the Dunn School of Pathology by Prof Susan Lea and Dr Errin Johnson, then relocated to the new Biochemistry Phase II building in 2021, COSMIC facilitates an exciting and diverse range of structural biology research at Oxford. It is open both to researchers (including external to Oxford University) and to industry.
COSMIC comprises of a fleet of high end instrumentation and computing for every stage of the cryo-EM workflow:
- FEI Talos 200 kV cryo-TEM with STEM detector for screening samples using negative staining and cryo-EM, as well as for cryo-tomography and cryo-STEM tomography
- FEI Arctica 200 kV cryo-TEM with FEI Falcon 4 direct electron detector for high throughput screening of cryo-grids and single particle analysis
- FEI Titan Krios 300 kV cryo-TEM with Gatan K3 direct electron detector + energy filter for high resolution single particle analysis, cryo-tomography and cryo-STEM tomography
- Zeiss Crossbeam 550 cryo-FIB-SEM for cryo-lamella production and cryo-volume SEM
- Three FEI Mark IV Vitrobots for preparing cryo-EM grids
- High performance computing for data storage and processing
- Dedicated staff to train and support users
Contacts
If you would like to find out more about doing cryo-EM at COSMIC, please get in touch:
- Prof Matthew Higgins, COSMIC Scientific Director: matthew.higgins@bioch.ox.ac.uk
- Dr Tanmay Bharat, COSMIC Deputy Director: tanmay.bharat@path.ox.ac.uk
- Rishi Matadeen, COSMIC microscope facility manager: rishi.matadeen@path.ox.ac.uk
- Teige Matthews-Palmer, cryo-EM support scientist: teige.matthews-palmer@path.ox.ac.uk
- Dr Joseph Caesar, COSMIC Computing Manager: joseph.caesar@path.ox.ac.uk
Recommended reading:
- Milne, J.L., et al. 2013. Cryo‐electron microscopy–a primer for the non‐microscopist. The FEBS journal, 280(1), pp.28-45.
- Bai, X.C., et al. 2015. How cryo-EM is revolutionizing structural biology. Trends in biochemical sciences, 40(1), pp.49-57.
- Fernandez-Leiro, R. and Scheres, S.H., 2016. Unravelling biological macromolecules with cryo-electron microscopy. Nature, 537(7620), pp.339-346.
- Higgins, M.K. and Lea, S.M., 2017. On the state of crystallography at the dawn of the electron microscopy revolution. Current Opinion in Structural Biology, 46, pp.95-101.
- Wu, M. and Lander, G.C., 2020. Present and emerging methodologies in cryo-EM single particle analysis. Biophysical Journal.
- Drulyte, I., Johnson, R.M., Hesketh, E.L., Hurdiss, D.L., Scarff, C.A., Porav, S.A., Ranson, N.A., Muench, S.P. and Thompson, R.F., 2018. Approaches to altering particle distributions in cryo-electron microscopy sample preparation. Acta Crystallographica Section D: Structural Biology, 74(6), pp.560-571.
- Lučić, V., et al. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol, 202(3), pp.407-419.
- Melia, C.E. and Bharat, T.A., 2018. Locating macromolecules and determining structures inside bacterial cells using electron cryotomography. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1866(9), pp.973-981.
- Böhning, J. and Bharat, T.A., 2020. Towards high-throughput in situ structural biology using electron cryotomography. Progress in Biophysics and Molecular Biology.
Recent publications from COSMIC:
- Deme JC, Johnson S, Vickery O, Aron A, Monkhouse H, Griffiths T, James RH, Berks BC, Coulton JW, Stansfeld PJ, Lea SM. 2020. Structures of the stator complex that drives rotation of the bacterial flagellum. Nature Microbiology. 5(12): 1553-1564.
- Gérard, S.F., Hall, B.S., Zaki, A.M., Corfield, K.A., Mayerhofer, P.U., Costa, C., Whelligan, D.K., Biggin, P.C., Simmonds, R.E. and Higgins, M.K., 2020. Structure of the inhibited state of the Sec translocon. Molecular Cell, 79(3), pp.406-415.
- von Kügelgen, A., Tang, H., Hardy, G.G., Kureisaite-Ciziene, D., Brun, Y.V., Stansfeld, P.J., Robinson, C.V. and Bharat, T.A., 2020. In Situ structure of an intact lipopolysaccharide-bound bacterial surface layer. Cell, 180(2), pp.348-358.
- Johnson, S., Fong, Y.H., Deme, J.C., Furlong, E.J., Kuhlen, L. and Lea, S.M., 2020. Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation. Nature Microbiology, pp.1-10.
- Kuhlen, L., Johnson, S., Zeitler, A., Bäurle, S., Deme, J.C., Caesar, J.J., Debo, R., Fisher, J., Wagner, S. and Lea, S.M., 2020. The substrate specificity switch FlhB assembles onto the export gate to regulate type three secretion. Nature communications, 11(1), pp.1-10.
- Tarafder, A.K., von Kügelgen, A., Mellul, A.J., Schulze, U., Aarts, D.G. and Bharat, T.A., 2020. Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria. Proceedings of the National Academy of Sciences, 117(9), pp.4724-4731.
- Lauber, F., Deme, J.C., Lea, S.M. and Berks, B.C., 2018. Type 9 secretion system structures reveal a new protein transport mechanism. Nature, 564(7734), p.77.